(The R code for Barnard’s exact test is at the end of the article, and you could also just download it from here, or from github)
About Barnard’s exact test
About half a year ago, I was studying various statistical methods to employ on contingency tables. I came across a promising method for 2×2 contingency tables called “Barnard’s exact test“. Barnard’s test is a non-parametric alternative to Fisher’s exact test which can be more powerful (for 2×2 tables) but is also more time-consuming to compute (References can be found in the Wikipedia article on the subject).
The test was first published by George Alfred Barnard (1945) (link to the original paper in Nature). Mehta and Senchaudhuri (2003) explain why Barnard’s test can be more powerful than Fisher’s under certain conditions:
When comparing Fisher’s and Barnard’s exact tests, the loss of power due to the greater discreteness of the Fisher statistic is somewhat offset by the requirement that Barnard’s exact test must maximize over all possible p-values, by choice of the nuisance parameter, π. For 2 × 2 tables the loss of power due to the discreteness dominates over the loss of power due to the maximization, resulting in greater power for Barnard’s exact test. But as the number of rows and columns of the observed table increase, the maximizing factor will tend to dominate, and Fisher’s exact test will achieve greater power than Barnard’s.
About the R implementation of Barnard’s exact test
After finding about Barnard’s test I was sad to discover that (at the time) there had been no R implementation of it. But last week, I received a surprising e-mail with good news. The sender, Peter Calhoun, currently a graduate student at the University of Florida, had implemented the algorithm in R. Peter had found my posting on the R mailing list (from almost half a year ago) and was so kind as to share with me (and the rest of the R community) his R code for computing Barnard’s exact test. Here is some of what Peter wrote to me about his code:
On a side note, I believe there are more efficient codes than this one. For example, I’ve seen codes in Matlab that run faster and display nicer-looking graphs. However, this code will still provide accurate results and a plot that gives the p-value based on the nuisance parameter. I did not come up with the idea of this code, I simply translated Matlab code into R, occasionally using different methods to get the same result. The code was translated from:
Trujillo-Ortiz, A., R. Hernandez-Walls, A. Castro-Perez, L. Rodriguez-Cardozo. Probability Test. A MATLAB file. URL
http://www.mathworks.com/matlabcentral/fileexchange/loadFile.do?objectId=6198
My goal was to make this test accessible to everyone. Although there are many ways to run this test through Matlab, I hadn’t seen any code to implement this test in R. I hope it is useful for you, and if you have any questions or ways to improve this code, please contact me at [email protected]
Using the R function for Barnard’s exact test
# source("https://www.r-statistics.com/wp-content/uploads/2010/02/Barnard.R.txt") # downloading the code from the website
# Loading the function
source("https://www.r-statistics.com/wp-content/uploads/2012/01/source_https.r.txt") # Making sure we can source code from github
source_https("https://raw.github.com/talgalili/R-code-snippets/master/Barnard.R")
# Using the function
#Examples:
Convictions <-matrix(c(2, 10, 15, 3), nrow = 2, dimnames = list(c("Dizygotic", "Monozygotic"), c("Convicted", "Not convicted")))
fisher.test(Convictions, alternative = "less")
Barnard(Convictions)
And here is the output
> #Examples:
> Convictions <-matrix(c(2, 10, 15, 3), nrow = 2, dimnames = list(c("Dizygotic", "Monozygotic"), c("Convicted", "Not convicted")))
> fisher.test(Convictions, alternative = "less")
Fisher's Exact Test for Count Data
data: Convictions
p-value = 0.0004652
alternative hypothesis: true odds ratio is less than 1
95 percent confidence interval:
0.0000000 0.2849601
sample estimates:
odds ratio
0.04693661
> Barnard(Convictions)
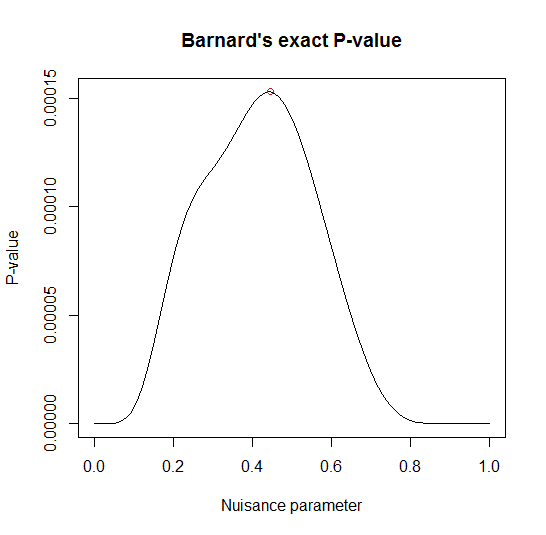
2x2 matrix Barnard's exact test: 100 13x19 tables were evaluated
-----------------------------------------------------------
Wald statistic = 3.6099
Nuisance parameter = 0.44446
p-values: 1-tailed = 0.00015285 2-tailed = 0.00030569
-----------------------------------------------------------
Final note: I would like to thank Peter Calhoun again for sharing his code with the rest of us - Thanks Peter!
Update (21.04.2010): In case you are facing a table with structural zeros (that is, missing values in the table), the package aylmer might be able to help you (it offers a generalization of Fisher's exact test)
Update (16.06.2011): A more updated (and faster) code is now available on the post (thanks goes, again, to Peter Calhoun). The post is updated with a newer example. The old R code for the function can still be found here.
Update (12.04.2012): I just found out that there is a new R package (from 2011-11-21) that also implements Barnard's test. You can find it here.
Thank you for this. In clinical trials, I would dare say that there are too many times when Fisher’s exact test is used when Bernard’s is more appropriate because of the assumptions of fixed margins.
My pleasure.
And thanks for sharing your experience about the overuse of fisher exact test.
Thanks so much for this. Just finished analysis, and the code worked great. Among the programs I have access to, this is the ONLY place I was able to find a resource for calculating Barnard’s Exact Test.
Good things happen when you open the source…
Hi Andrew – thank you for sharing in your experience 🙂
All the best,
Tal
Hi Tal,
Very helpful codes for 2 by 2 contigency table.
I have couple of questions,
1. Does Barnard’s method offer P value based on two-tailed test ? Within Fisher Exact’s method, the P value for two-tailed test could be different substantially from twice of 1-tailed test when the counts are small, unlike the symmetric distribution cases.
2. matrix (c(6,81,0,29)) gives p=0.120034 vs matrix (0,29,6,81) gives p=0.1198472
Is that what we expect from Barnard method?
Thanks,
Joey
Same for me. Thank you very much for the code! 🙂
Tal, Thanks so much for the code!
I’m trying to figure out how to get directionality of positive results out of a cont. table test, i.e in which tail is a given result found.
Is there a good way to do this? Which cells in a 2×2 table would I compare?
Hi Dan,
It depends on how your table is structured – but you could simply look at the table and describe the direction observed (the Barnard’s exact test wouldn’t do this for you)
¡Mil gracias!
Thank you so much for this lovely present.
You’re welcome 🙂
In case anyone still views this blog, I published the R code under the Exact package. The newest code has more options and unconditional tests, is more efficient, and can calculate the power under the binomial models.
im trying to use the exact package for this matrix:
raw_matrix = structure(c(343, 11, 28825, 2014),
.Dim = c(2L, 2L), .Dimnames = list(
c(“NotH3K”, “H3K”), c(“CNS”, “Random”)))
using the command Exact(raw_matrix) it doesnt work
giving error like this:
Error: cannot allocate vector of size 42.0 Gb
could you tell me what im doing wrong here?
Cheers for this, works nicely. The only free way of doing the Barnard’s text I’ve found yet!
For those reading this who don’t know R, it’s easy enough to use. I used 3.0 for windows 7 64 bit. The free package can be downloaded (~50mB) and installed in a few minutes. Once installed & you’re connected to the web, boot it up, then go to the packages menu, click “install packages”. Follow instructions. I used the UK (London) mirror, and then selected ‘Barnard’. Once you’ve installed and loaded the “Barnard” package, you can run it by using the command as described in the link in the 12/04/12 update.
In brief, it’s “barnardw.test(n1, n2, n3, n4, dp = 0.001, verbose = FALSE)” then hit enter.
The numbers of you 2×2 grid are:
n1, n2,
n3, n4.
These settings worked fine for me. If you need more detailed settings, you’ll be bright enough to find the info in that link above!
All in all, 5 test took me a total of about 20 mins to run, and that includes the time to download, install, and learn how to boot up R, install and load my package, and run the calculations. Not bad at all!
Thanks again guys!
I cannot thank you enough for generously sharing the script. Without it I wouldn’t be able to move on to another stage of my study.
Hat off to you.
and anohter hat off to Peter Calhoun !!